|
|
Input sequence (single letter code)
Type or cut and paste your query sequence into the form. The amino acid sequence must be in the standard single letter code format. Spaces within the pasted sequence are permitted, however they will be removed with the remaining sequence treated as a single contiguous chain.
It is recommended that you enter the sequence as a single-letter string like this: IMWRNAKRQSDRFYDEDVFINGEGLEPEQDTRGVDNAHMVTNH
DISOPRED2 will not accept sequences with lengths greater than 1000 amino acids. Better predictions are likely to be obtained by splitting the protein into domains, using a domain boundary recognition method such as DomPred, and submitting each separately.
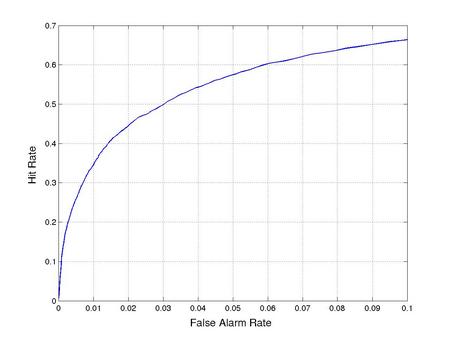
Disordered regions represent less than 5% of the residues in the solved crystal structures used for training DISOPRED2. As a result, trivially predicting order for all residues achieves high accuracy without providing informative classifications. It is therefore necessary to set a false positive rate threshold for the classifier. The receiver-operating characteristic (ROC) curve shows the fraction of disordered residues recovered for a particular false hit rate. For example, assuming the user wishes to recover at least half of the disordered residues, the false positive rate threshold should be set at 3%. This also results in a relatively high precision for the residues classed as disordered (table 1).
Figure 1: ROC curves for disorder prediction

Table 1: Precision/recall characteristics for false positive rate thresholds
| False Positive Rate | Precision | Recall | False Positive Rate | Precision | Recall | |
|---|---|---|---|---|---|---|
| 0.01 | 0.49 | 0.35 | 0.06 | 0.22 | 0.60 | |
| 0.02 | 0.39 | 0.44 | 0.07 | 0.20 | 0.62 | |
| 0.03 | 0.32 | 0.50 | 0.08 | 0.18 | 0.64 | |
| 0.04 | 0.28 | 0.54 | 0.09 | 0.17 | 0.65 | |
| 0.05 | 0.25 | 0.57 | 0.10 | 0.16 | 0.66 |
Secondary structure assignments from the PSIPRED prediction method can optionally be included in the results. More information on PSIPRED can be found at www.psipred.net. Regular secondary structure elements (i.e. helix and sheet) can be disordered as a result of flexibility in the intervening regions of globally disordered proteins. However, local disorder should not generally overlap with segments that have a high probability of forming helix or strand. These disordered regions are likely to be composed of highly flexible residues on the protein surface.
The results are sent in plain ASCII text format but include links to secondary structure graphics and disorder profile plots.
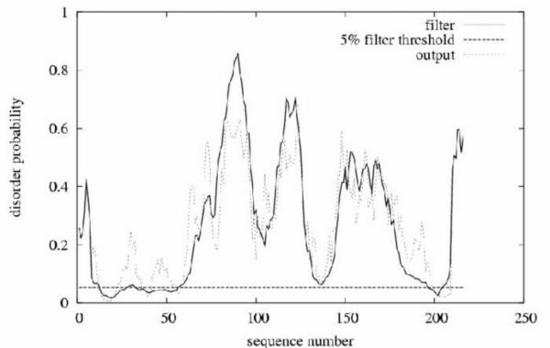
Example prediction for the intracellular loop of the membrane protein gliotactin from Drosophila . Gliotactin was classed as a completely unstructured protein in the CASP5 experiment. The plot shows position in the sequence against probability of being disordered. In total, 157 of the 216 residues are classed as disordered at the default threshold. The horizontal line is the order/disorder threshold for the default false positive rate of 5\%. The 'filter' curve represents the outputs from DISOPRED2 and the 'output' curve the outputs from a linear SVM classifier (DISOPREDsvm). The outputs from DISOPREDsvm are included to indicate shorter, low confidence predictions of disorder.
DISOPRED initially runs a PSI-BLAST search over a filtered sequence database. The position-specific scoring matrix at the final iteration of PSI-BLAST is used to generate inputs to the classifier. More details can be found at DISOPRED home. The user can choose to receive PSI-BLAST log files, however these files may be extremely large (~10 Mb) and it is requested that this option is only checked if absolutely necessary.
Please, insert a valid email address to receive the results, which will be returned as soon as they are available. This will usually be a few minutes but may be longer depending on server load. DISOPRED2 is free for academic use but access is forbidden for users with commercial email addresses. Enquiries for commercial use should be sent to: dtj@cs.ucl.ac.uk.
The short name you supply with the sequence will be included in the subject line of the email containing the results. This is useful for identifying each sequence if you intend to submit multiple jobs to the server.
Note that, at present, the DISOPRED server only has limited capacity. We plan to move the service to a high capacity server, but until then, we ask that you limit your usage to no more than 20 requests per day, and no more than 5 requests per hour. If you require a larger number of predictions then please get in touch to discuss your requirements.